Computes tests for each of the two factors and optionally for their interaction using likelihood ratio tests. p-values will not be identical to the usual method of moments ANOVA tests but will be similar.
cen2way(y1, y2, fac1, fac2, LOG = TRUE, interact = TRUE)Arguments
- y1
The column of data values plus detection limits
- y2
The column of indicators, where 1 (or
TRUE) indicates a detection limit in they1column, and 0 (orFALSE) indicates a detected value iny1.- fac1
The first grouping or factor variable. Can be either a text or numeric value indicating the group assignment.
- fac2
The second grouping or factor variable. Can be either a text or numeric value indicating the group assignment.
- LOG
A logical variable indicating whether natural logs are to be taken of the 'y1' column data. Default is TRUE.
- interact
A logical variable indicating whether to compute an interaction term between the two variables. Default is TRUE. #' @keywords two-way two-factor factorial ANOVA analysis of variance censored
Value
Q-Q plots of residuals. Likelihood ratio test statistics ("chisquare"), degrees of freedom ("df") and p-values (pval) for two factors and optionally the interaction. Data on the underlying models, including AIC and R2 are also provided.
Details
Tests are computed using Maximum Likelihood Estimation. When a gaussian distribution model is used (LOG=FALSE) modeled values may fall below zero, producing unreal p-values (often lower than they should be). Because of this, testing in log units is preferable and is the default unless you are transforming the y values prior to running the function (such as taking cube roots to approximate a gamma distribution). The 'delta.lr0x2' stat output is the -2loglikehood for the test of the model versus an intercept-only model.
References
Helsel, D.R., 2011. Statistics for Censored Environmental Data using Minitab and R, 2nd ed. John Wiley & Sons, USA, N.J. Millard, S.P., 2013. EnvStats: An R Package for Environmental Statistics. Springer-Verlag, New York.
See also
Examples
data(Gales_Creek)
Gales_Creek$Period <- c(rep("early", 35), rep("middle", 12), rep("late", 16))
with(Gales_Creek,cen2way(TCr, CrND, Season, Period))
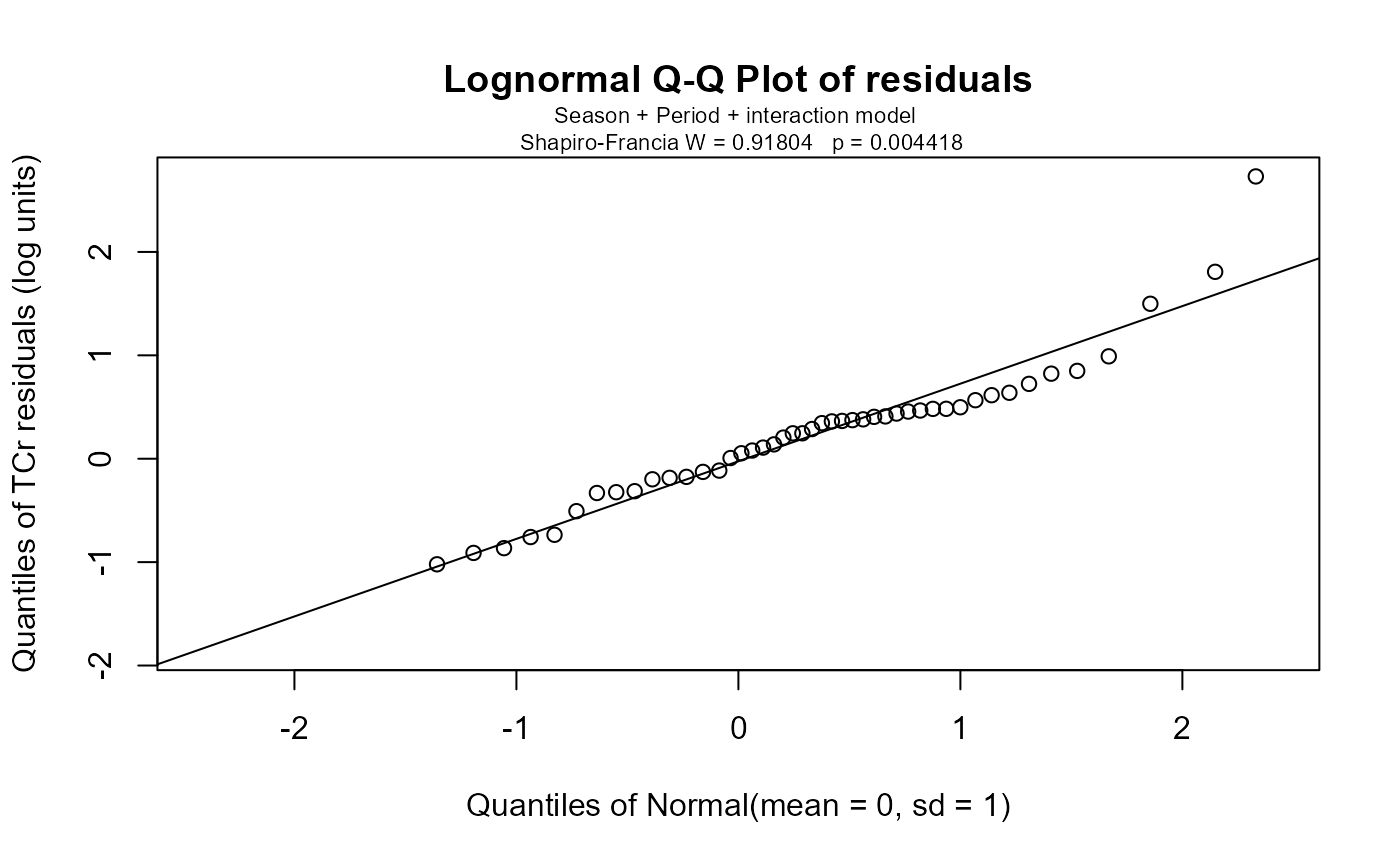 #> Two-way Fixed Effects ANOVA by MLE Likelihood Ratio Tests for Censored Data
#> ln(TCr) modeled by Factor 1: Season and Factor 2: Period
#> ANOVA Table with interaction
#> FACTORS chisquare df pval
#> Season 21.5100 1 3.520e-06
#> Period 5.5840 2 6.130e-02
#> interaction 0.8491 2 6.541e-01
#>
#> MODELS loglikelihood delta.lr0x2 AIC BIC
#> Season only -68.20914 24.700 147.4183 157.2127
#> Period only -76.16994 8.783 165.3399 177.2932
#> both factors -65.84173 29.440 142.6835 152.4779
#> both + interaction -65.41720 30.290 145.8344 159.9466
#> Rescaled_Loglik_R2
#> 0.3516
#> 0.1411
#> 0.4047
#> 0.4138
#> Two-way Fixed Effects ANOVA by MLE Likelihood Ratio Tests for Censored Data
#> ln(TCr) modeled by Factor 1: Season and Factor 2: Period
#> ANOVA Table with interaction
#> FACTORS chisquare df pval
#> Season 21.5100 1 3.520e-06
#> Period 5.5840 2 6.130e-02
#> interaction 0.8491 2 6.541e-01
#>
#> MODELS loglikelihood delta.lr0x2 AIC BIC
#> Season only -68.20914 24.700 147.4183 157.2127
#> Period only -76.16994 8.783 165.3399 177.2932
#> both factors -65.84173 29.440 142.6835 152.4779
#> both + interaction -65.41720 30.290 145.8344 159.9466
#> Rescaled_Loglik_R2
#> 0.3516
#> 0.1411
#> 0.4047
#> 0.4138